You can find the most updated list of publications here:
6.
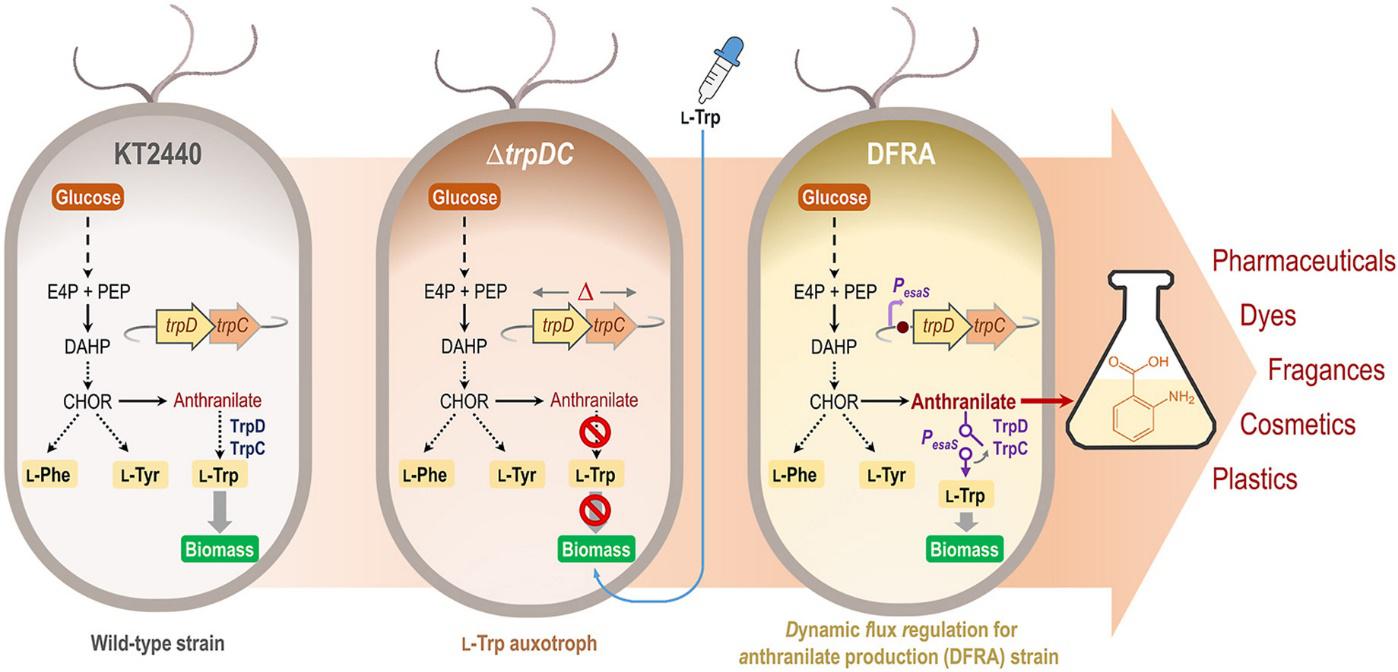
Fernández-Cabezón, L., Rosich i Bosch, B., Kozaeva, E., Gurdo, N., and Nikel, P. I. 1. (2022) Dynamic flux regulation for high-titer anthranilate production by plasmid-free, conditionally-auxotrophic strains of Pseudomonas putida. Metabolic Engineering, DOI: https://doi.org/10.1016/j.ymben.2022.05.008
Lorena and Berta developed synthetic gene circuits based on quorum sensing elements, and used the toolset to boost the production of anthranilate, an aromatic compound highly relevant for industrial applications, by playing with conditional auxotrophies and cell densities.
Visit Publication5.
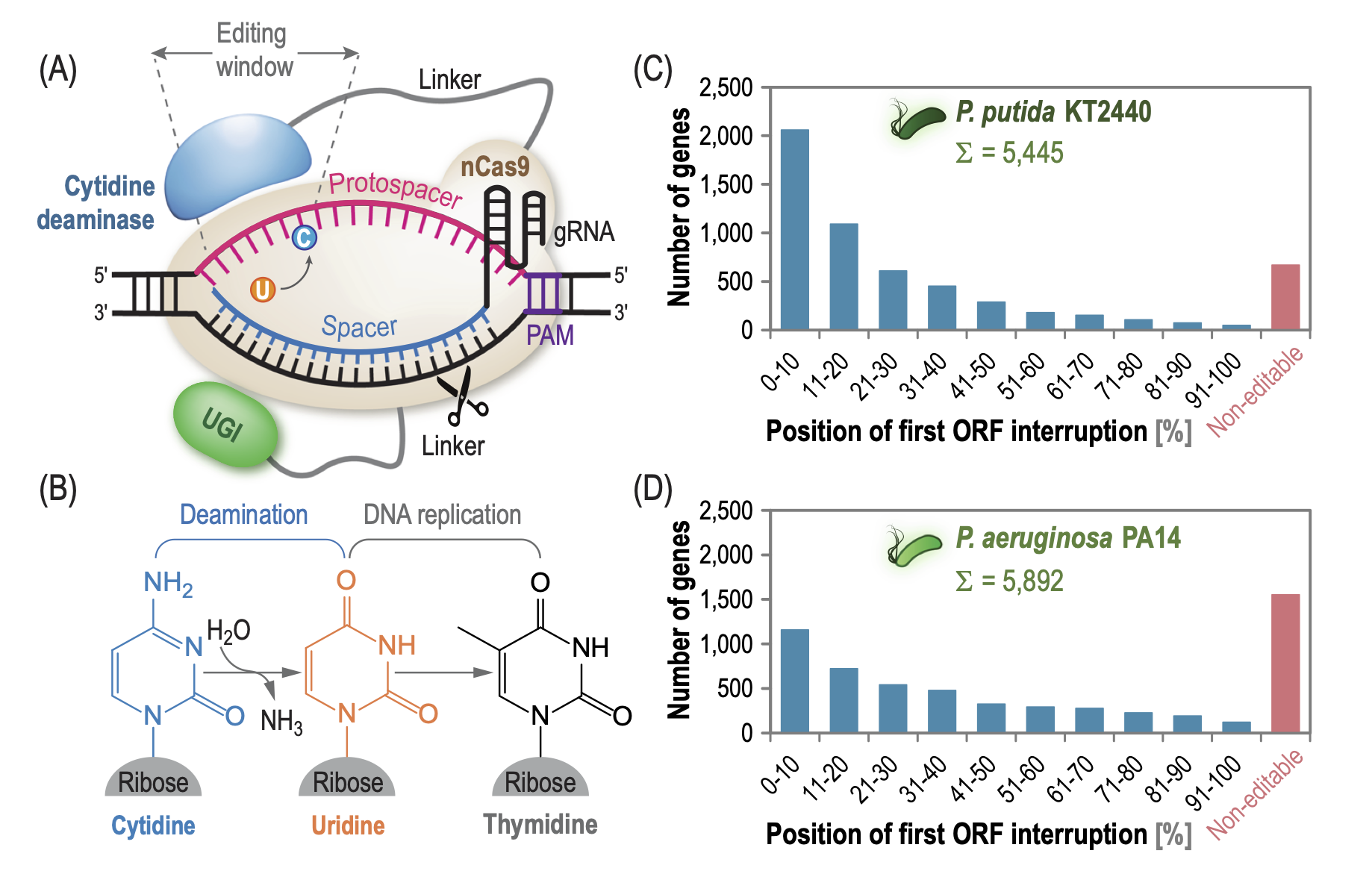
Volke, D. C., Martino, R. A., Kozaeva, E., Smania, A. M., and Nikel, P. I. (2022) Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9-assisted, multiplex cytidine base-editing. Nature Communications, DOI: https://doi.org/10.1038/s41467-022-30780-z
In this research article, Daniel and Ekaterina, together with Román, report a fast and efficient CRISPR/nCas9-based toolbox for engineering the genome of Pseudomonas and other Gram-negative bacteria, and they provide different examples of deeply engineered strains obtained either by single-step or sequential genome-editing efforts.
Visit Publication4.
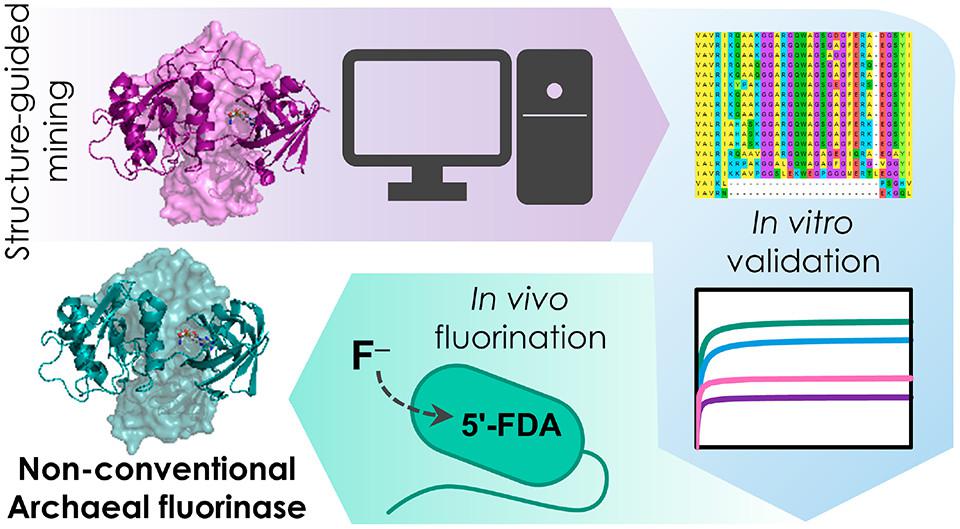
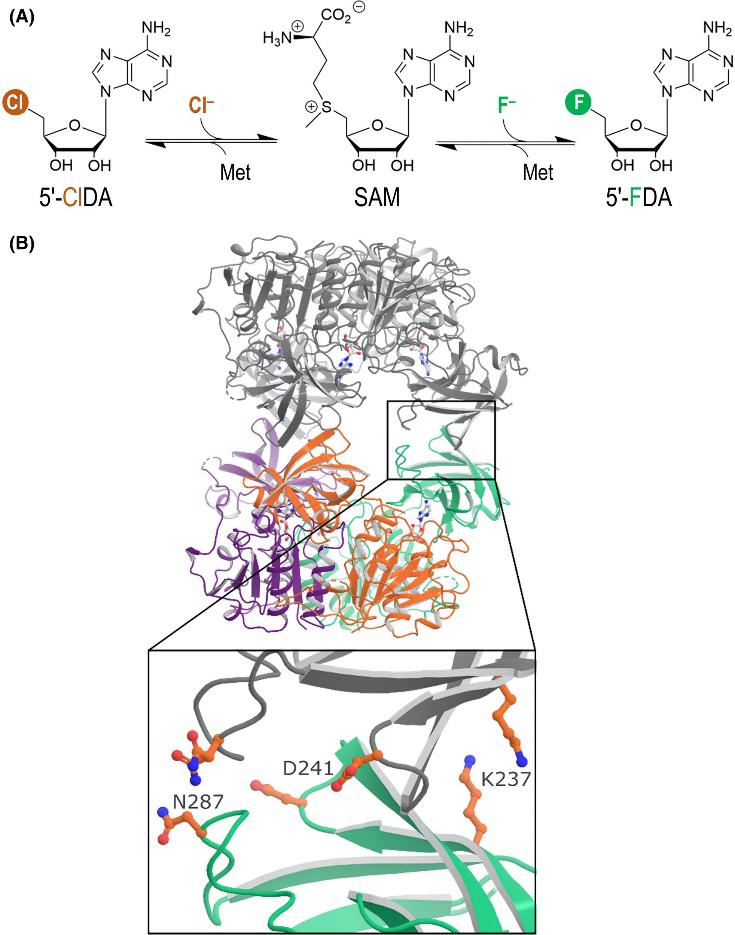
Pardo, I., Bednar, D., Calero, P., Volke, D. C., Damborský, J., and Nikel, P. I. (2022) A nonconventional Archaeal fluorinase identified by in silico mining for enhanced fluorine biocatalysis. ACS Catalysis, DOI: https://doi.org/10.1021/acscatal.2c01184
Isabel characterized novel fluorinases, identified by in silico mining as part of a collaboration with Prof. J. Damborský’s laboratory. One of them, a putative ‘chlorinase’ from an Archaeal species, turned to be the best performing fluorinase up to date!
Visit Publication3.
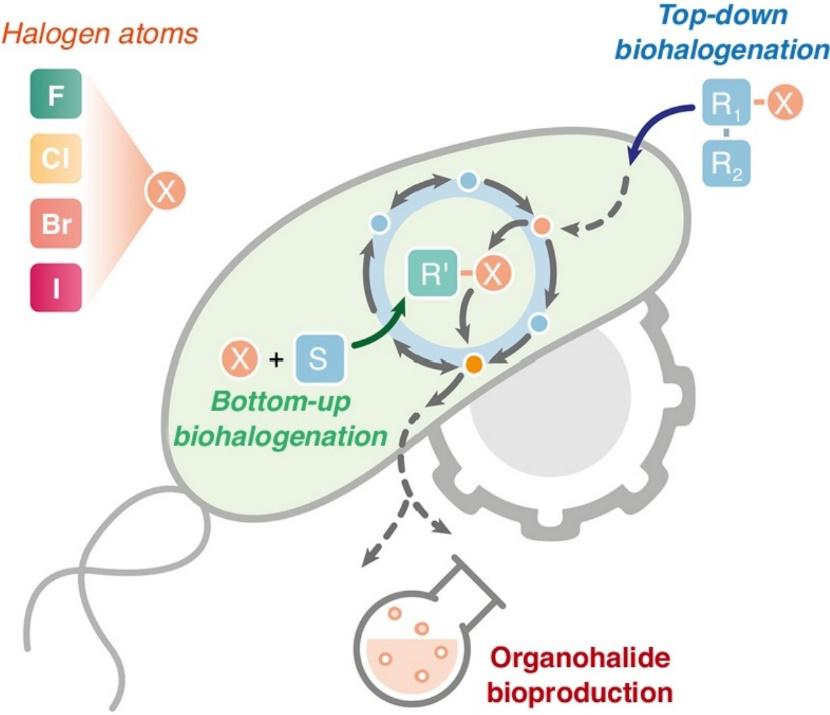
Cros, A., Alfaro-Espinoza, G., De Maria, A., Wirth, N. T., and Nikel, P. I. (2022) Synthetic metabolism for biohalogenation. Current Opinion in Biotechnology, DOI: https://doi.org/10.1016/j.copbio.2021.11.009.
In this review article, Antonin, Alberto, Nico Wirth and Pablo discuss the importance of exploiting cell factories and synthetic biology for production of organohalides, as well as bringing to the table different strategies to achieve this goal.
Visit Publication2.
Kittilä, T., Calero, P., Fredslund, F., Lowe, P. T., Tezé, D., Nieto-Domínguez, M., O’Hagan, D., Nikel, P. I., and Welner, D. H. 5. (2022) Oligomerization engineering of the fluorinase enzyme leads to an active trimer that supports synthesis of fluorometabolites in vitro. Microbial Biotechnology, DOI: https://doi.org/10.1111/1751-7915.14009
In this collaboration paper with Tiia and led by D. Welner, we explored the structure and mechanism of action of one of our favorite enzyme, the fluorinase, and demonstrate that a trimeric version (rather than its hexameric form), is still active.
Visit Publication1.
Arce-Rodríguez, A., Pankratz, D., Preusse, M., Nikel, P. I., and Häussler, S. (2022) Dual effect: High NADH levels contribute to efflux-mediated antibiotic resistance but drive lethality mediated by reactive oxygen species. mBio, 6. DOI: https://doi.org/10.1128/mbio.02434-21
In this article, the importance of the redox balance in bacteria for antibiotic resistance was assessed. Cells need NADH to support different resistance mechanisms but, at the same time, this redox ‘pressure’ contributes to the accumulation of ROS—a fine-tune balance is thus of crucial importance towards fighting the antibiotic resistance crisis.
Visit Publication