You can find the most updated list of publications here:
11.
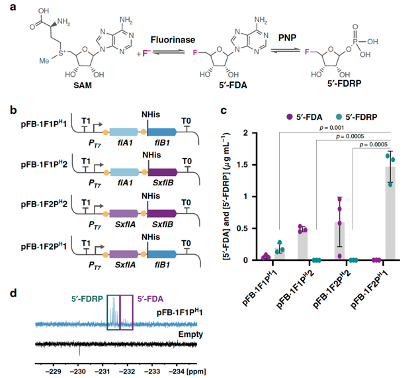
Calero, P., Volke, D. C., Lowe, P. T., Gotfredsen, G. H., O’Hagan, D., and Nikel, P. I. (2020) A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida. Nature Communications, 11 : 5045. DOI: 10.1038/s41467-020-18813-x
A first-case example of how to engineer cell factories for the biosynthesis of fluorometabolites. Patricia has used a riboswitch to produce fluorinases in vivo, where fluoride is both an inducer of gene expression and the substrate for the synthetic biofluorination module.
Visit Publication10.
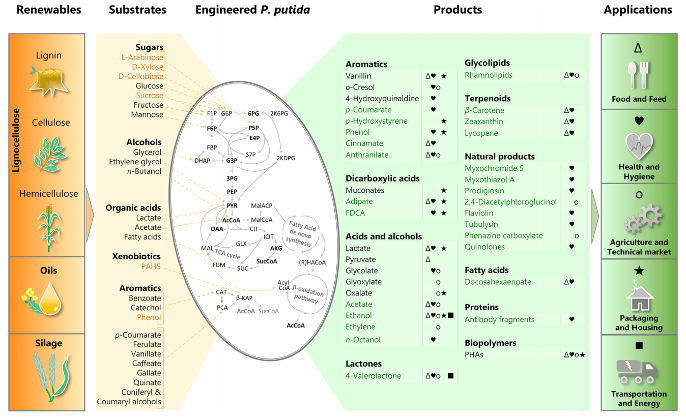
Weimer, A., Kohlstedt, M., Volke, D. C., Nikel, P. I. and Wittmann, C. (2020) Industrial biotechnology of Pseudomonas putida: advances and prospects. Applied Microbiology and Biotechnology, 104: DOI: 10.1007/s00253-020-10811-9
In this mini-review, we summarize the recent advances in genetic engineering, systems and synthetic biology and practical applications of P. putida as a cell factory.
Visit Publication9.
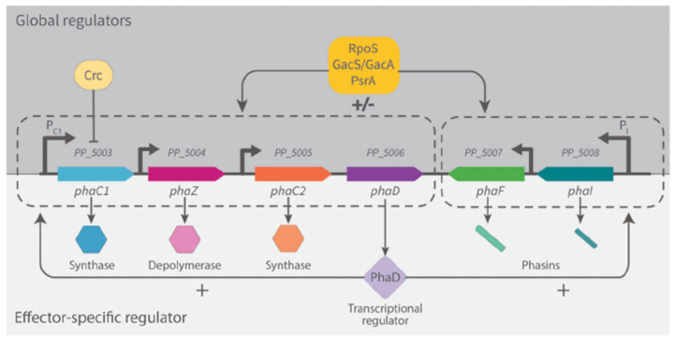
Mezzina, M. P., Manoli, M. T., Prieto, M. A., and Nikel, P. I.(2020) Engineering native and synthetic pathways in Pseudomonas putida for the production of tailored polyhydroxyalkanoates. Biotechnology Journal, DOI: 10.1002/biot.202000165
Mariela and Pablo, together with our expert collaborators in CSIC, have put together a thorough revision of different engineering strategies for PHA production in our favorite bug Pseudomonas putida. The article focuses on the biosynthesis of new-to-Nature polymers, which include rare chemical elements in their structure.
Visit Publication8.
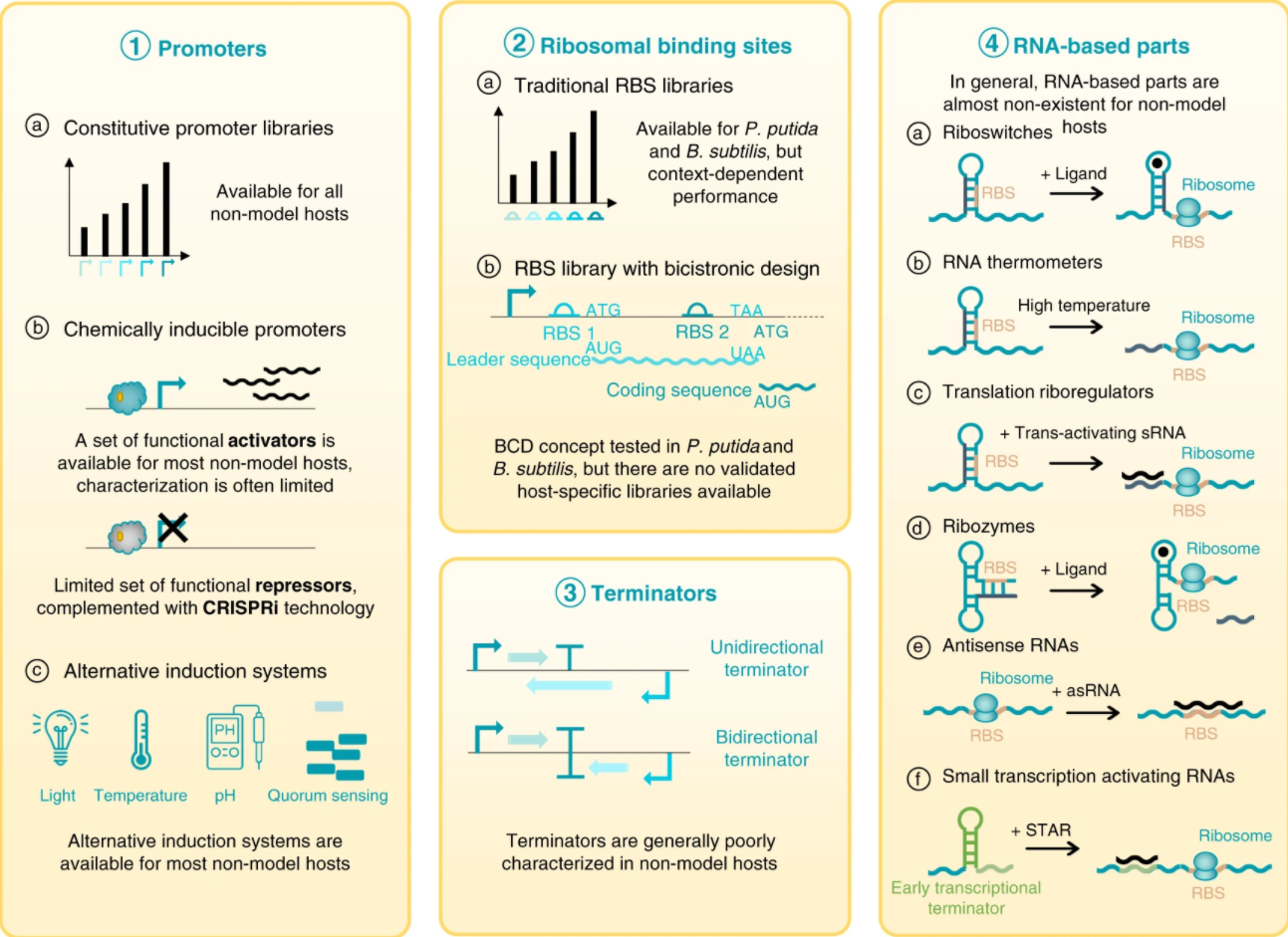
Lammens, E. M., Nikel, P. I., and Lavigne, R. (2020) Exploring the synthetic biology potential of bacteriophages for engineering non-model bacteria. Nature Communications, 11: 5294. DOI: 10.1038/s41467-020-19124-x
What are the SynBio parts available for engineering non-traditional microbial hosts? We explore this question in this review article together with our collaborators in Leuven, Belgium. Bacteriophages are a treasure trove of devices that can be used for gene and genome engineering of bacterial platforms!
Visit Publication7.
Wurtzel, E. T., Vickers, C. E., Hanson, A. D., Millar, A. H., Cooper, M., Voss-Fels, K. P., Nikel, P. I., and Erb T. J. (2020) Revolutionizing agriculture with synthetic biology. Nature Plants, 5: 1207-1210. DOI: 10.1038/s41477-019-0539-0
How can SynBio mediate a revolution in plant research – especially for agricultural applications? Our two-cents on the matter in this opinion article where we discuss approaches, currently validated in microorganism, could be potentially transferred into plants.
Visit Publication6.
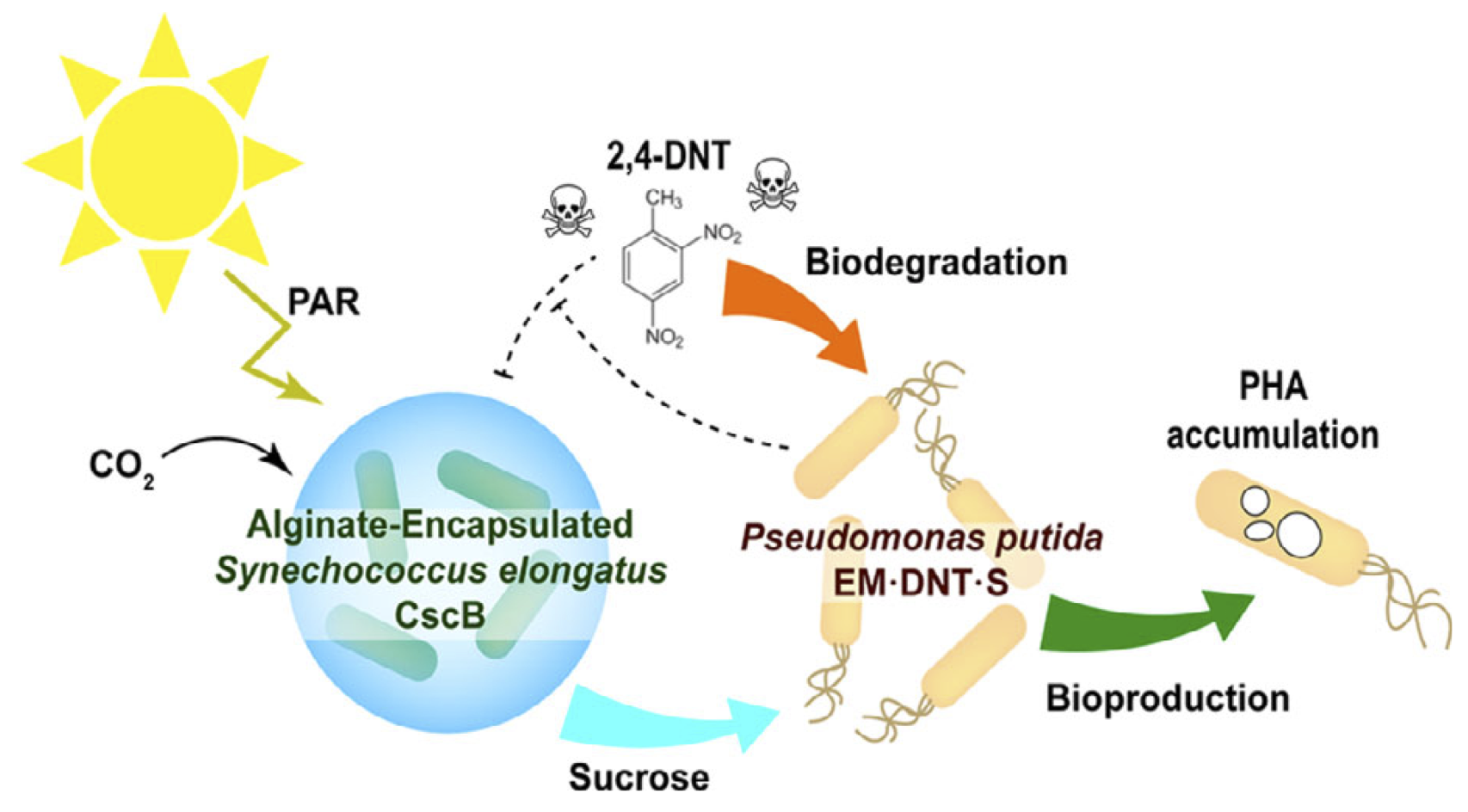
Fedeson, D. T., Saake, P., Calero, P., Nikel, P. I., and Ducat, D.C. (2020) Biotransformation of 2,4-dinitrotoluene in a phototrophic co-culture of engineered Synechococcus elongatus and Pseudomonas putida. Microbial Biotechnology, DOI: 10.1111/1751-7915.13544
A synthetic microbial consortium composed by two engineered microorganisms, the photosynthetic cyanobacterium Synechococcus elongatus PCC 7942 and the heterotrophic bacterium Pseudomonas putida EM173, performs degradation of the pollutant 2,4-dinitrotoluene using CO2 and light as the only inputs in the system.
Visit Publication5.
Wirth, N. T., and Nikel, P. I. (2020) Engineering reduced-genome strains of Pseudomonas putida for product valorization. In Minimal Cells: Design, Construction, Biotechnological Applications. DOI: 10.1007/978-3-030-31897-0_3
A book chapter describing the state-of-the-art of P. putida genome engineering, focusing on reduced-genome strains with enhanced traits for production, product valorization, and development of cell factories.
Visit Publication4.
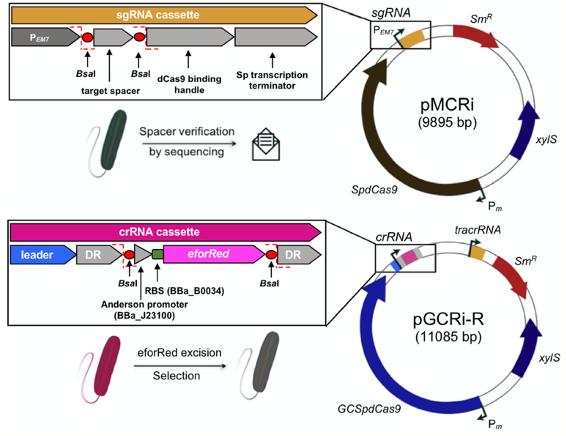
Batianis, C., Kozaeva, E., Damalas, S. G., Martín‐Pascual, M., Volke, D. C., Nikel, P. I., and Martins dos Santos, V. A. P. (2020) An expanded CRISPRi toolbox for tunable control of gene expression in Pseudomonas putida. Microbial Biotechnology, 13: 368-385. DOI: 10.1111/1751-7915.13533
Broadening the SynBio toolbox for P. putida! In this case, we describe a protocol for single-plasmid CRISPR-interference (CRISPRi) for down-regulation of one or multiple gene targets in Pseudomonas developed by Ekaterina.
Visit Publication3.
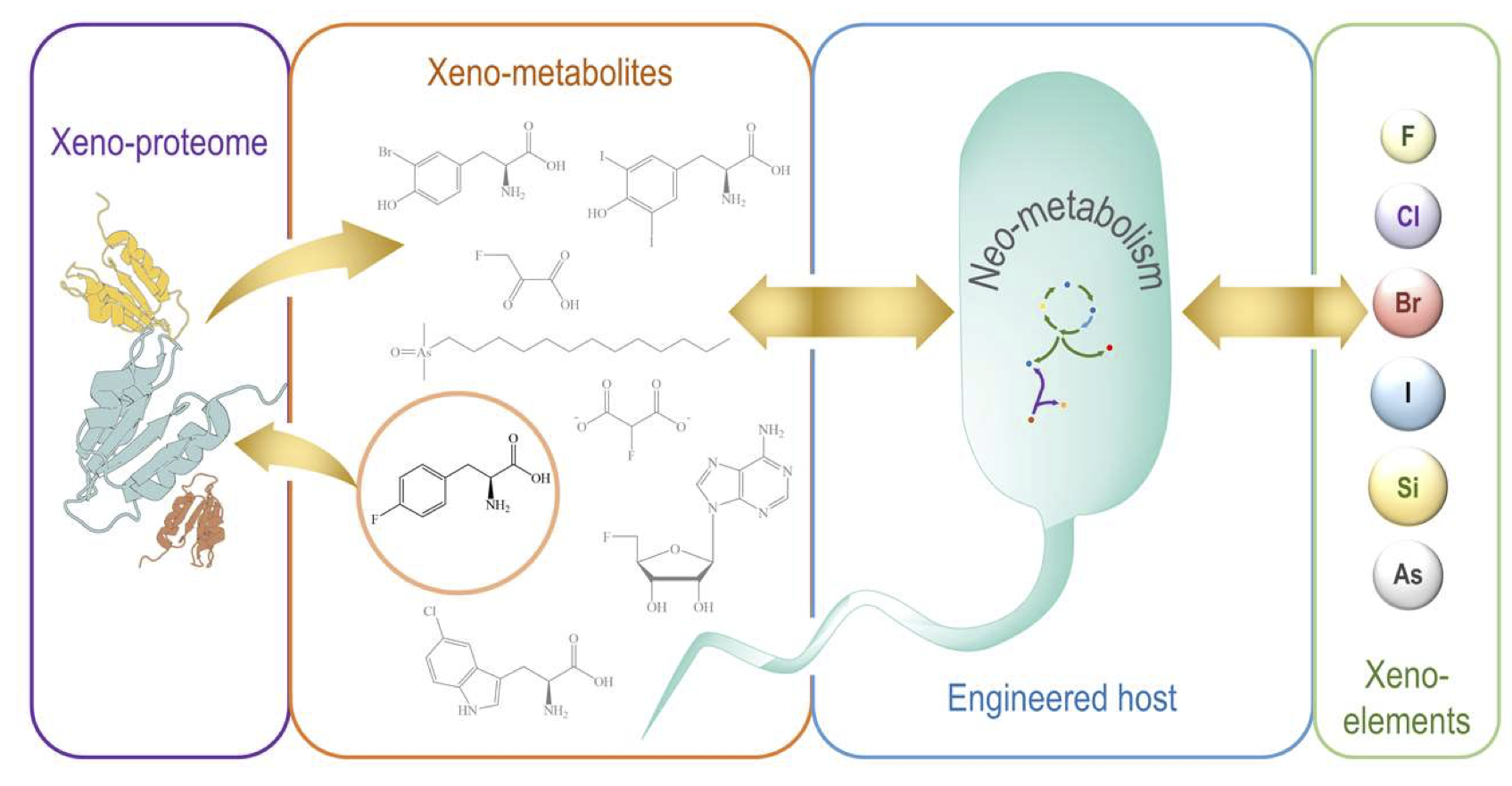
Nieto-Domínguez, M. and Nikel, P. I. (2020) Intersecting xenobiology and neometabolism to bring novel chemistries to life. ChemBioChem, 3. DOI: 10.1002/cbic.202000091
Manuel and Pablo explore the possibility of incorporating non-biological elements into the biochemistry of microorganisms and review different examples where this effort has been accomplished. SynBio strategies to create novel (neo) metabolism are discussed with a focus on fluorine incorporation into organic molecules.
Visit Publication2.
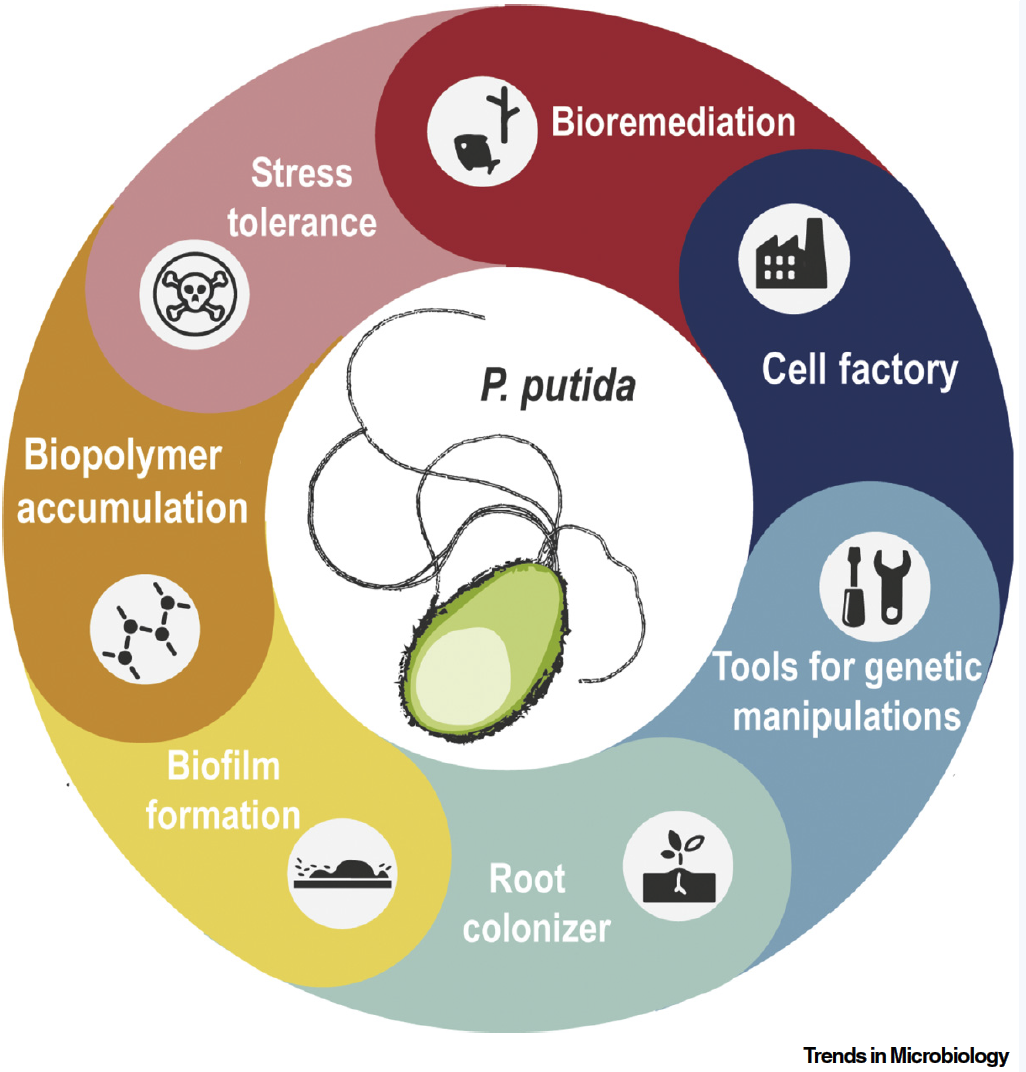
Volke, D. C., Calero, P., and Nikel, P. I. (2020) Pseudomonas putida. Trends in Microbiology, 28, 512-513. DOI: 10.1016/j.tim.2020.02.015
A glimpse on what makes Pseudomonas putida such a special host, including applications and highlighting milestones of the continuously expanding P. putida-related research.
Visit Publication1.
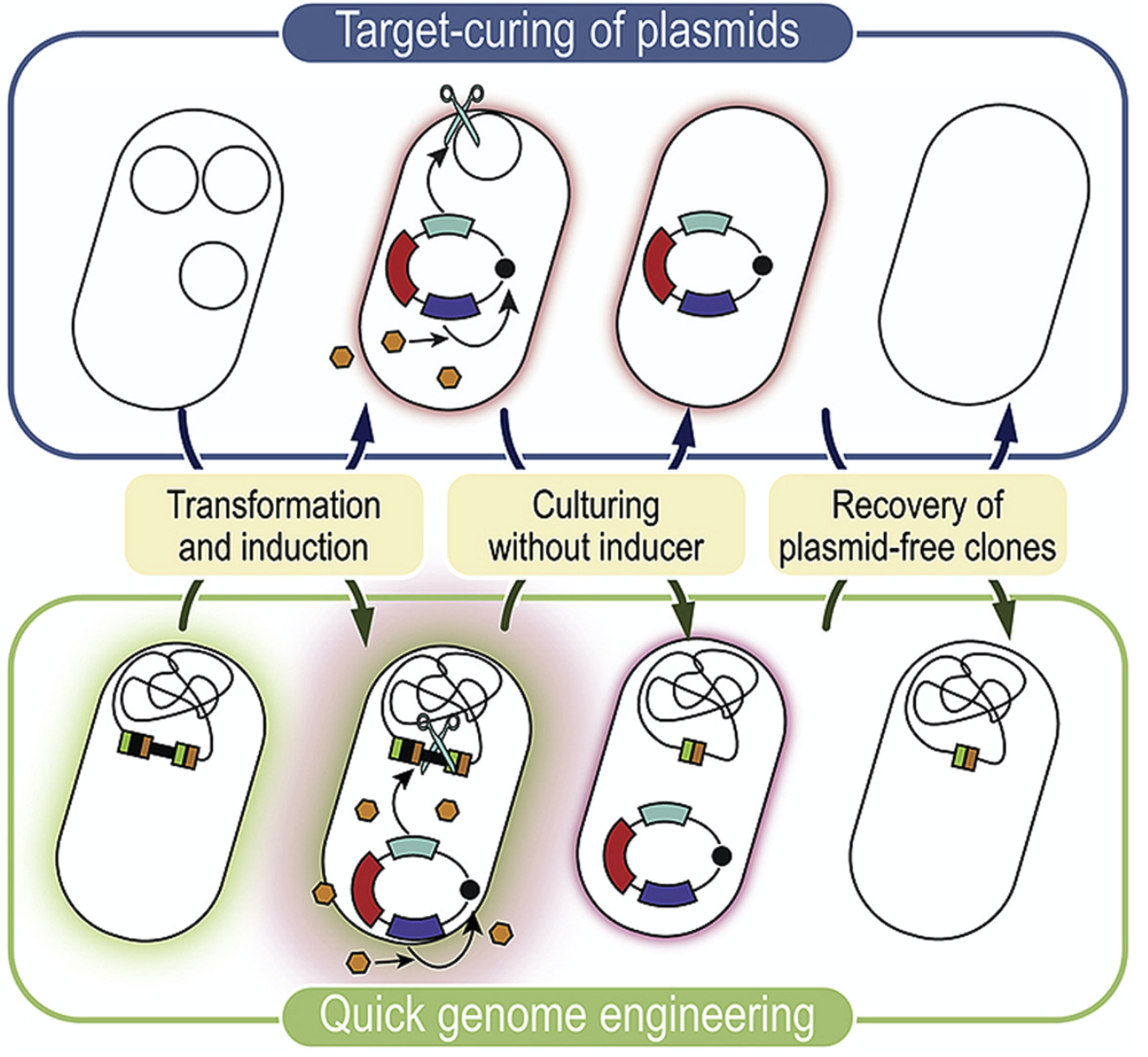
Volke, D. C., Friis, L., Wirth, N. T., Turlin, J., and Nikel, P. I. (2020) Synthetic control of plasmid replication enables target- and self-curing of vectors and expedites genome engineering of Pseudomonas putida. Metabolic Engineering Communications, 10: e00126. DOI: 10.1016/j.mec.2020.e00126
We describe new methods for curing plasmids in P. putida using vectors that can target themselves via the expression of an endonuclease to speed up engineering efforts in Pseudomonas. Most plasmids can be cured from >95% of the whole bacterial population after an overnight cultivation!
Visit Publication